The intestinal microbiota plays a key role in maintaining the metabolic homeostasis of the host. The absence or dysregulation of the intestinal microbiota heavily affects dietary polysaccharides digestion and lipid absorption, leading to dysfunction of liver and adipose tissue, and thus resulting in a series of metabolic diseases such as obesity, type 2 diabetes, and cardiovascular diseases. Although the close relationship between gut microbiota and host metabolism has been reported by a large number of studies, the underlying mechanism by which gut microbiota regulates host metabolism remains to be elucidated.

On August 25, 2023, Dr. Wang Yuhao, the principal investigator of The First Affiliated Hospital of Zhejiang University School of Medicine/Zhejiang University Institute of Translational Medicine//National Center for Basic Sciences, teamed up with Dr. Lora Hooper, professor of the University of Texas Southwestern Medical Center at Dallas in the USA, published a research article entitled “The gut microbiota reprograms intestinal lipid metabolism through long non-coding RNA Snhg9” in Science journal. This study unveiled a new mechanism by which the intestinal microbiota regulates host lipid metabolism through long non-coding RNA (lncRNA).
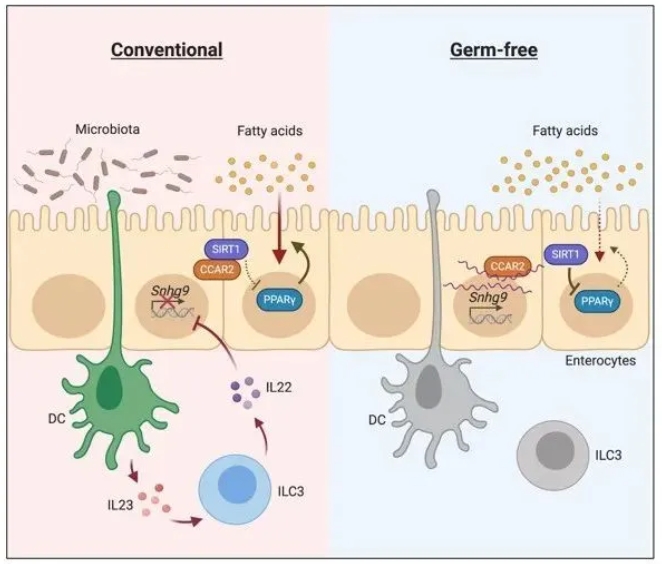
Studies on germ-free mice, which do not have any bacteria in the body, have repeatedly found that the absence of microbiota can lead to lowered body fat percentage and resistance to high-fat diet-induced obesity, however, the mechanisms behind this phenomenon are not fully understood. By comparing the transcriptomes of intestinal epithelial cells between germ-free mice and conventionally raised mice, researchers from Dr. Yuhao Wang’s team found that the transcription level of a lncRNA, named Snhg9, was significantly increased in germ-free mice, indicating that the expression of this lncRNA is regulated by the abundance of gut microbiota. Because intestinal epithelial cells are the main cell type for dietary long-chain fatty acids absorption and initial processing, this finding raised a hypothesis that the high expression of Snhg9 in intestinal epithelial cells could be one of the reasons for abnormal lipid absorption in germ-free mice.
Based on this hypothesis, the research team conducted a series of experiments and uncovered the essential role of Snhg9 in regulating intestinal lipid absorption. Specifically, Snhg9 directly binds to CCAR2 (cell cycle and apoptosis regulator 2) - an inhibitor of deacetylase SIRT1 (sirtuin 1), and dissociates CCAR2 from SIRT1, leading to increased deacetylation activity of SIRT1. SIRT1 is a major repressor of core lipid metabolic transcription factor PPARγ, thus increased SIRT1 activity results in repressed expression of PPARγ. Therefore, when the researchers overexpressed Snhg9 in mice intestinal epithelial cells, these mice exhibited suppressed lipid absorption and reduced body fat percentage, proving the close relationship between Snhg9 and intestinal lipid metabolism.
Since the expression of Snhg9 is suppressed by the microbiota, the researchers further explored the mechanism by which the microbiota suppresses the expression of Snhg9. They found that the microbiota did not directly inhibit the expression of Snhg9 in intestinal epithelial cells, but through a subepithelial relay involving myeloid cells and type 3 innate lymphoid cells (ILC3) instead. Therefore, the immune system is also crucial for microbiota regulation of host metabolism.
In summary, this study is among the first to unveil the key role of lncRNA Snhg9 in regulating intestinal lipid metabolism, reveal the signaling pathway behind microbiota-suppressed Snhg9 expression, and elucidate the mechanism by which Snhg9 inhibits lipid absorption. This study also suggests new strategies for clinical treatment of metabolic diseases by targeting the microbiota or Snhg9. It is worth noting that this study is another major discovery made by the team of Dr. Yuhao Wang and Dr. Lora Hooper following the discovery of the key regulatory proteins NFIL3 (Science 2017) and HDAC3 (Science 2019) in microbial-host interaction.
Source: Zhejiang University Institute of Translational Medicine


